Epidrug Repurposing: Discovering New Faces of Old Acquaintances in Cancer Therapy
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7708379/
Figure 1

Overview of the epigenetic landscape. Different compaction levels of chromatin are depicted, from naked DNA to the metaphasic chromosome. (A) Two dimers of H2A-H2B and a tetramer of H3-H4 histones are required for nucleosome assembly, the chromatin’s basic packaging unit (B) DNA methylation is a process carried out by DMNTs in CpG dinucleotides, particularly on CpG islands. This dynamic epigenetic mark can be reversed by enzymatic conversion. (C) Histone acetylation is performed on lysine residues by HAT enzyme complexes. In contrast, histone lysine deacetylation is carried out by HDACs enzyme complexes. (D) Histone lysine methylation is carried out by HMT complexes. Lysines can be processively methylated from mono to di and trimethylation.
후생유전학적 풍경의 개요.
Naked DNA에서 metaphasic 염색체에 이르기까지 염색질의 다양한 압축 수준이 표시됩니다.
(A) H2A-H2B의 두 이량체와 H3-H4 히스톤의 사량체가 뉴클레오솜 조립에 필요하며, 염색질의 기본 패키징 단위
(B) DNA 메틸화는 CpG 디뉴클레오티드, 특히 CpG 섬에서 DMNT에 의해 수행되는 과정입니다.
이 역동적인 후성 유전적 표시는 효소적 전환에 의해 역전될 수 있습니다.
(C) 히스톤 아세틸화는 HAT 효소 복합체에 의해 라이신 잔기에서 수행됩니다.
대조적으로, 히스톤 라이신 탈아세틸화는 HDAC 효소 복합체에 의해 수행됩니다.
(D)히스톤 라이신 메틸화는 HMT 복합체에 의해 수행됩니다.
라이신은 모노에서 디 및 트리메틸화로 과정적으로 메틸화될 수 있습니다.
Figure 2

Epigenetic alterations in cancer cells. In non-neoplasic cells, CpG islands of tumor suppressor gene promoters are generally unmethylated and acetylated, resulting in transcriptional activation and expression. In contrast, non-coding regions and repetitive elements are hypermethylated, ensuring chromosome stability. Gene bodies are normally methylated, enhancing transcription. Neoplasic cells are characterized by global hypomethylation and local CpG island hypermethylation, especially at tumor suppressor gene promoters, resulting in aberrant transcription and genomic instability.
암세포의 후성유전학적 변화.
비종양 세포에서 종양 억제 유전자 프로모터의 CpG 섬은 일반적으로 메틸화되지 않고 아세틸화되어 전사 활성화 및 발현을 초래합니다.
대조적으로, 비암호화 영역과 반복 요소는 과메틸화되어 염색체 안정성을 보장합니다.
유전자 본체는 일반적으로 메틸화되어 전사를 향상시킵니다.
신생물 세포는 특히 종양 억제 유전자 프로모터에서 전체적인 저메틸화 및 국소 CpG 섬 과메틸화를 특징으로 하여 비정상적인 전사 및 게놈 불안정성을 초래합니다.
Figure 3
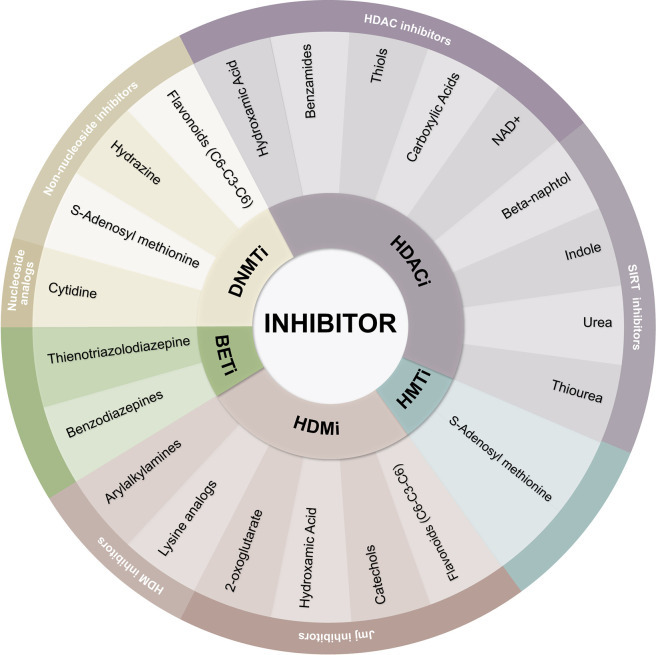
Classification of epigenetic inhibitors. Epigenetic inhibitors are classified as DNMTi, HDACi, HMTi, HDMI, and BETi. The chemical nature of each inhibitor defines the affinity of its targets.
후성 유전적 억제제의 분류.
후성유전학적 억제제는 DNMTi, HDACi, HMTi, HDMI 및 BETi로 분류됩니다.
각 억제제의 화학적 성질은 표적의 친화성을 정의합니다
Table 2
Overview of epigenetic inhibitors currently in clinical trials for cancer therapies.
InhibitorMechanism of ActionFunctional Molecule or Chemical GroupExamplesCASClinical TrialsPhase StudiesConditionsIIIIIIIV
| DNMTi | Nucleoside analogs: Cytidine analogs incorporate into DNA instead of cytidine, covalently linking the enzyme and leading to DNMT degradation | Cytidine | Azacytidine | MDS, CML, AML, glioma, prostate cancer, pancreatic cancer, ovarian cancer, metastatic melanoma. | |||||
| Decitabine | CML, AML, MDS, prostate cancer, thyroid cancer. | ||||||||
| Guadecitabine | AML, MDS, HCC, CMML, ovarian cancer, urothelial carcinoma, colorectal cancer, peritoneal cancer | ||||||||
| 5-fluoro-2’-deoxycytidine | AML, MDS, Head and Neck Neoplasms, Lung Neoplasms, Urinary Bladder Neoplasms, Breast Neoplasms | ||||||||
| 4’-thio-2’-deoxycytidine | Currently establishing the safety, tolerability, and MTD in patients with refractory solid tumors. | ||||||||
| Non-nucleoside inhibitors either block the DNMTs enzyme catalytic site, interact with enzyme recognition of target sequences or are SAM cofactor competitors. | S-Adenosyl methionine | Sinefungin | NA | ||||||
| Hydrazine | Hydralazine | ovarian cancer, cervical cancer, refractory solid tumors, breast cancer. | |||||||
| Flavonoids (C6-C3-C6) | Epigallocatechin-3-gallate | Adenocarcinoma of the prostate, head and neck cancer, colon cancer, pancreatic cancer, breast cancer, lung cancer, bladder cancer, colorectal cancer, prostate cancer. | |||||||
| HDACi | HDACi are molecules capable of Zinc trapping that bind to the zinc-containing catalytic domain of HDACs and supress their deacetylase enzymatic activity | Hydroxamic Acid | Vorinostat | Rhabdomyosarcoma, Leiomyosarcoma, Lymphoma, melanoma, Lung carcinoma, lung cancer, head and neck cancer, leukemia, breast cancer, MDS, ovarian cancer, glioblastoma, pancreatic cancer, breast cancer. | |||||
| Trichostatin A | Relapsed or Refractory Hematologic Malignancies | ||||||||
| Belinostat | MDS, Non-Hodgkin lymphona, mantle cell lymphoma, diffuse large B-cell lymphoma, breast cancer, ovarian cancer, lung cancer, glioblastoma, AML, ATLL, bladder cancer, liver cancer, | ||||||||
| Panobinostat | AML, MDS, lung cancer, gliosarcoma, prostate cancer, multiple myeloma, CMML, breast cancer, pancreatic cancer. | ||||||||
| dacinostat | NA | ||||||||
| givinostat | chronic lymphocytic leukemia, multiple myeloma, hodgkin’s lymphoma. | ||||||||
| Benzamides | Entinostat | breast cancer, prostate adenocarcinoma, renal cell carcinoma, lymphoma, MDS, melanoma, lung cancer, AML, colorectal cancer, pancreatic cancer | |||||||
| mocetinostat | urothelial carcinoma, Hodgkin lymphoma, Head and Neck cancer, MDS, lung cancer, melanoma. | ||||||||
| Thiols | Romidepsin | T cell lymphoma, glioma, multiple myeloma, CTCL, leukemia, astrocytoma, pancreatic cancer, lung cancer, thyroid cancer, prostate cancer, male breast cancer, renal cancer, bladder cancer. | |||||||
| Carboxylic Acids | Valproic acid | AML, MDS, Head and Neck cancer, SCC, glioma, bladder cancer, sarcoma, glioblastoma, leukemia, breast cancer, lung cancer. | |||||||
| Butyric Acid | schyzofrenic disorders | ||||||||
| Phenylbutiric Acid | colon cancer, leukemia, gastric cancer, MDS. | ||||||||
| Pivanex | melanoma, lung cancer, leukemia. | ||||||||
| SIRTi are small molecules, many of them recently discovered by cell-based screening assays, with multiple inhibition mechanisms including reactivity with chemical intermediates, non-competitive inhibition with substrate and uncompetitive inhibition with NAD+. | NAD+ | Nicotin | NA | ||||||
| beta-naphtol | sirtinol | NA | |||||||
| splitomicin | NA | ||||||||
| salermide | NA | ||||||||
| cambinol | NA | ||||||||
| indole | EX-527 | Endometriosis | |||||||
| oxyndole | NA | ||||||||
| urea | suramin | lung cancer, breast cancer, adrenocortical carcinoma, renal cancer, prostate cancer, bladder cancer, multiple myeloma, head and neck cancer. | |||||||
| thiourea | tenovin | NA | |||||||
| HMTi | HKMTi are SAM like molecules and molecules that directly inhibits the enzyme S-adenosyl-L-homocysteine hydrolase or interact with the cofactor binding pocket of KMTs | S-Adenosyl methionine | Sinefungin | NA | |||||
| EPZ004777 | NA | ||||||||
| EPZ-5676 | AML, MDS, leukemia | ||||||||
| EPZ004777 | NA | ||||||||
| Valemetostat | leukemia, lymphoma, prostate cancer, renal cancer. | ||||||||
| tazemetostat | B cell lymphoma, prostate cancer, mesothelioma, Non Hodgkin lymphoma, tissue sarcoma, Bladder cancer, sinonasal carcinoma, follicular lymphoma. | ||||||||
| Most HRMT inhibitors are molecules which occupy and inhibit the SAM pocket, the substrate pocket, or both. | S-Adenosyl methionine | GSK3326595 | neoplasms | ||||||
| HDMi | HDM inhbitors are molecules that inhibit monomine oxidases family of enzymes or that are substrate mimics (lysine analogs). | Arylalkylamines | Phenelzine | breast cancer, prostate cancer. | |||||
| Tranylcypromine | AML, MDS | ||||||||
| Pargyline | NA | ||||||||
| Lysine analogs | propylhydrazine | NA | |||||||
| JmjC inhibitors are derivates of 2OG, hydroxamic acids, catechols and flavonoids. | 2-oxoglutarate | N-oxalylglicine | NA | ||||||
| Hydroxamic Acid | Methylstat | NA | |||||||
| Catechols | Hematoxylin | NA | |||||||
| Caffeic acid | esophagus cancer | ||||||||
| Flavonoids (C6-C3-C6) | Myricetin | NA | |||||||
| Baicalein | Influenza | ||||||||
| Epigallocatechin-3-gallate | Adenocarcinoma of the prostate, head and neck cancer, colon cancer, pancreatic cancer, breast cancer, lung cancer, bladder cancer, colorectal cancer, prostate cancer. | ||||||||
| BETi | BET inhibitors are derivates of benzodiazepines that take up the hydrophobic región of BET enzymes which binds acetylated lysines. | Thienotriazolodiazepines | JQ1 | NA | |||||
| CPI-203 | NA | ||||||||
| OTX015 | AML, glioblastoma, breast cancer, lung cancer, prostate cancer. | ||||||||
| Benzodiazepines | CPI-0610 | Myeloma, lymphoma, leukemia, MDS. | |||||||
| Molibresib | lymphoma, NUT carcinoma, |
Table 4
Current DNMTi and DNMT-HDAC dual inhibitors repurposed drugs with applications in cancer therapy [*modified from Moreira-Silva et al. (9)].
ClassCompoundFirst indicationEpigenetic targetDrug-target interactionCancer model/New indicationKey features in mechanismReferences
| Section 1. DNMT inhibitors | |||||||
| Non-nucleoside analogs | Hydralazine | Anti-hypertensor | DNMT1 | Four high-affinity interaction points with DNMT1 through the residues Lys 162 and Arg 240 within the enzyme active site. | T‐Cell Leukemia cells | Increases LFA-1 expression inhibits T Cells ERK pathway phosphorylation, decreases DNMT enzyme activity, and decreases DNMT1 and DNMT3A protein levels. Reduces de novo methylation due to greater affinity to hemi methylated substrates (target of DNMT1),. | (156, 166) |
| Breast Cancer cells | In vivo induces DNA demethylation and increases expression of ER as well as RARb, p12 and p16 in vitro. | (167) | |||||
| Bladder Cervical Cancer cells | (157) | ||||||
| Prostate Cancer cells | Increases apoptosis, inhibits RGFR pathway, thus induces cell cycle arrest. Decreases DNMT1, DNMT3a and b protein levels. Upregulates p21 which decreases promoters DNA methylation and induces histone acetylation. | (156) | |||||
| Cervical Cancer cells | Induces APC expression, inhibits cell growth, induces cell cycle arrest and apoptosis. Promotes DNA demethylation. | (158) | |||||
| Disulfiram (DSF) | Alcohol aversive | DNMT1 | DSF could interfere with the catalytic activity of DNMT1 by reacting with a citosine ring via thiol group of catalytic site of DNMT1. | Prostate Cancer cells | Reduces global 5mC content, through inhibition of DNMT1 activity on hemimethylated substrates. Decreases methylation in APC and RARB gene promoters, thus increasing re-expression. Inhibits growth and clonogenic survival of prostate cancer cell lines. | (70) | |
| Procainamide | Cardiac arrythmias | DNMT1 | Partially competitive inhibitor of DNMT1 that interacts with the binding pocket of the enzyme | Prostate Cancer cells | Promotes GSTP1 CpG island hypomethylation, thus induces GSTP1 re-expression in LNCaP cells in vitro and in in vivo assays | (168) | |
| Breast Cancer cells | Induces DNA demethylation increases expression of ER RARb; also induces re-expression of p12 and p16 (in vitro). | (169) | |||||
| Colon Cancer cells | Greatly reduces affinity for hemi-methylated DNA and SAM in catalysis, reduces global 5mC content, thus reduces gene-specific hypermethylation at promoter CpG islands. | (154) | |||||
| Non-small Cell Lung Cancer cells | Inhibits DNMT activity and decreases promoter demethylation of WIF-1, restoring WIF-1 expression, thus downregulating the Wnt pathway | (169) | |||||
| Procaine (PCA) | Anesthesic for spinal block | DNMT1, DNMT3A | Interacts with the binding pocket of the enzyme inhibiting catalytic activity (non-nucleoside), | Breast Cancer cells | Demethylates densely hypermethylated CpG islands, reduces 5mC DNA content by 40%, restoring gene expression of RARβ2, and has growth-inhibitory effects, causing mitotic arrest | (153) | |
| Gastric Cancer cells | Inhibits DNMT1 and 3A activity through molecular docking in the catalytic binding site, disrupting the binding of DNMT to DNA. Reduces proliferation, induces apoptosis, and restores expression of CDKN2A and RARb | (152) | |||||
| Hepatocellular Carcinoma cells | DNA demethylation and silenced gene reactivation of p16, HAI-2/PB, and NQO1. Promotes cell cycle arrest and reduces viability. Also shown significant reduction in tumor volume in vivo. | (170) | |||||
| Non-small Cell Lung Cancer cells | Inhibits DNMT activity, causing promoter demethylation of WIF-1, thus restores WIF-1 expression and downregulation of Wnt pathway | (169) | |||||
| Antibiotic | Nanaomycin A | Anthracycline antibiotic | DNMT3B | Interaction with active site of DNMT3B in specific a.a. (Glu697 Arg731 Arg733) of enzyme binding pocket, thus promoting a molecular docking in DNMT3B that inhibits enzymatic activity | Colon Cancer cells | Decreases DNMT1, 3A, 3B expression. Inhibits DNMT3B activity promoting reactivation of RASSF1A. Reduces cell proliferation and viability | (164) |
| Lung Cancer cells | |||||||
| Bone marrow cells | |||||||
| Mithramycin A (MMA) | Hypercalcemia drug, and antineoplastic agent | DNMT1 | Possibly interferes with DNMT1 binding at the CpG region in TSG promoters through binding DNMT1 protein or might be a form a complex between MMA, DNMT1 and double-stranded DNA |
Lung Cancer cells | Inhibits DNMT1 activity and decreases protein level. Decreases CpG methylation on SLIT2 and TIMP-3 promoters, inducing re-expression. Inhibits invasor pehotype thus prevents metastasis | (163) | |
| Polyphenol | Chlorogenic acid | Natural Compound (not approved) | DNMT1 | Increases SAH formation inhibiting DNA methylation through COMT mechanism (non-competitive), | Breast Cancer cells | Inhibits DNMT1 activity, reduces methylation of the promoter region of the RARb gene | (150) |
| Harmine | Natural Compound (not approved) | DNMT1 | Not described | Myeloid Leukemia cells | Decreases DNMT1 gene expression, induces p15 promoter demethylation. Also decreases proliferation and promotes cell cycle arrest in G0/G1 phase | (149) | |
| Laccaic acid A (LCA) | Natural Compound (not approved) | DNMT1 | DNA-competitive DNMT inhibitor through competition for the oligonucleotide substrate | Breast Cancer cells | Inhibits directly DNMT1, also have effects on DNMT3A, 3B inhibition, and reactivates genes silenced by promoter methylation (CEACAM5, DHRS3, RGS16) | (151) | |
| Mahanine | Natural Compound (not approved) | DNMT1, DNMT3B | Induces proteasomal degradation of DNMT1 and DNMT3B | Prostate Cancer cells | Inhibits DNMT activity, increases expression of RASSF1A and inhibits cyclin D1. Induces proteosomal degradation on DNMT1 and DNMT3B through Akt inactivation, thus facilitates demethylation of RASSF1A promoter and increases its expression | (155, 171) | |
| Genistein | Natural compound; isoflavone | DNMT | Inhibits DNA methyltransferase activity in a substrate- and methyl donor–dependent manner | Esophageal Squamous Cell Carcinoma cells | Promotes reversed DNA hypermethylation and reactivation of RARbeta, p16INK4a and MGMT trhough demethylation of promoter genes. Also inhibites cell growth |
(172) | |
| Prostate Cancer cells | Induces DNA hypomethylation and reactivation of RARbeta | ||||||
| Breast Cancer cells | |||||||
| Renal Cancer cells | Inhibits DNMT1 activity, thus induces demethylation of BTG3 promoter and has antiproliferative effects through cell cycle arrest | (173) | |||||
| Peptide | Beta amiloid peptide | Component of Alzheimer’s senile plaque | DNMT | Drecreases SAM/SAHlevels, promoting a global demethylation, through redox-dependent control over methionine synthase and methylation |
Neuroblastoma cells | Soluble Aβ oligomers decreases intracellular glutathione levels by hampering cysteine uptake, followed by a global decrease in global DNA methylation | (174) |
| BCM7 | Natural Compound; food-derived peptide | DNMT | Decreases SAM/SAH levels, promoting decrease in cysteine levels, affecting redox status and methylation capacity of DNMTs | Neuroblastoma cells | Decreases cysteine absorption through opioid receptor activation. This reduction is followed by an increase of oxidized glutathione and an increase in DNA methylation | (175, 176) | |
| GM7 | Natural Compound; food-derived peptide | DNMT | Neuroblastoma cells | ||||
| Section 2. DNMT and HDAC Dual Inhibitors | |||||||
| Polyphenol | Parthenolide | Anti‐inflammatory (not approved) | HDAC1 and DNMT1 | Inhibits DNMT1possibly through alkylation of the proximal thiolate of Cys1226 of the catalytic domain by its -methylene lactone |
Colon Cancer cells | Inhibits HDAC activity by molecular docking, downregulates HIF-1alfa and inhibits NF-kB pathway | (177, 178) |
| Melanoma cells | Reduces MITF-M transcript level and HDAC1 and protein level. | (179) | |||||
| Breast Cancer cells | Induces proteasomal degradation of HDAC1, thus increasing global histone acetylation and p21/p53 expression and induces cell death. | (179–181) | |||||
| Thyroid Cancer cells | Down-regulates DNMT1 expression possibly associated with its SubG1 cell-cycle arrest. Promotes global DNA hypomethylation and reactivates HIN-1 gene trough demethylation of its promoter | (181) | |||||
| Myeloid Leukemia (AML) cells | Inhibits DNMT1 and decreases gene expression of DNMT1 and BF-kB pathway. Induces cell cycle arrest and interrupts the binding of Sp1 to DNMT1 promoter, thus reactivates tumor suppressor genes and inhibites HIF-α | (178, 181) | |||||
| Resveratrol (RVT/RSV) | Natural Compound polyphenol (not approved); | HDAC1 and DNMT | Fits into the binding pocket of HDAC’s though interaction with amino acids of the catalytic site and interacts with the zinc ion, disrupting HDAC-zinc dependent activity. | Hepatoblastoma cells | Antiproliferative effect on all cell lines; showed specific inhibition of HDACs and in turn a histone hyperacetylation in HepG2 cells. | (182) | |
| Breast Cancer cells | Decreases PRMT5 EZH2 ATP2A3 and HDAC2 expression, increasing H3ac and H3K27 marks; increases global level of H3K9ac and H3K27ac marks through increasing KAT2A/3B expression. Reduces the enrichment of H4R3me2s and H3K27me3; and increases activating histone marks (H3K9/27ac) within the proximal promoter region of BRCA1, p53, and p21 restoring its expression. | (183, 184) | |||||
| Thyroid Cancer cells | Decreases DNMT1 activity and demethylates CpG sites at promoters regions in CRABP2. | (185) | |||||
| EGCG | Natural Compound; thiol anti-inflammatory | DNMT and HDAC | Inhibitor of DNMT nuclear activity by direct interaction with a hydrophilic pocket of DNMT1 and catalytic binding site | Squamous Cell carcinoma cells | Induces reversal hypermethytlation in RARβ, MGMT, p16INK4a, and hMLH1 promoter genes, promotes inhibition of cell growth. | (186, 187) | |
| Skin Cancer cells | Increases the levels of acetylation of histone H3 and histone H4 lysine residues through HDAC inhibition, leading to the upregulation of Cip1/p21 and p16INK4a. | (188) | |||||
Table 5
Current HDACi repurposed drugs with applications in cancer therapy [*modified from Moreira-Silva et al. (9)].
ClassCompoundFirst indicationEpigenetic targetDrug-target interactionCancer model/New indicationKey features in mechanismReferences
| HDAC inhibitors | |||||||
| Acid | Valproate (VPA) | Antiepileptic | HDAC class I and HDAC2 | Inhibits HDAC class I activity by binding to the catalytic site and promotes proteasomal degradation of HDAC2. | Melanoma treatment | Potentiates karenitecin-induced apoptosis in multiple melanoma cell lines and on xenografted mice, however, fails to enhance chemotherapy effects on dacarbazine plus interferon-α-treated melanoma patients. | (197, 198) |
| Colon Cancer cells | Reduces relative HDAC2 mRNA expression, preventing cell colony formation and migration. | (199) | |||||
| Non-small Cell Lung Cancer cells | Increases major histocompatibility complex (MHC) class I chain-related protein A (MICA) expression and sensitizes cancer cells to γδ T-cell-mediated killing. | (200) | |||||
| Colon Cancer Tumor cells | Synergistically reduces viability of cancer cells in combination with mytomicin C. | (201) | |||||
| Ovarian Cancer cells | Upregulates WWOX and P27 genes and interferes with the cell cycle by promoting apoptosis and inhibiting cell proliferation, both in vitro and in vivo. | (202) | |||||
| Phenolic | Artemisin | Antimalarial | HDAC1, HDAC2 and HDAC6 | Not described | Breast Cancer cells | Inhibits cell proliferation, cell migration, invasion and induces apoptosis. Also inhibits HDAC 1, 2, 6 and up-regulates BRCA1, 2/Ras/ERα/ERβ/PR/Her expression | (203) |
| Ginseng | Nutraceutical (not approved) | HDAC | Not described | Lung Carcinoma cells | Inhibits HDAC activity, increases p21 expression and induces apoptosis. | (204) | |
| HC toxin | Natural Compound antiprotozoal (not approved); | HDAC | Not described | Breast Cancer cells | Inhibits cell proliferation and induces cell cycle arrest at G2/M and apoptosis in a dose-dependent manner. | (205) | |
| Neuroblastoma Cells | Induces cell cycle arrest and apoptosis, induces neuronal differentiation and inhibits invasive growth. Increases p-RB, p15, p16, p21, p27 expression, and reactivates the RB tumor suppressor pathway. Also induces H4 acetylation while inhibits HDAC activity | (206) | |||||
| Psammaplin A (PsA) | Enzimatic inhibitor Bromotyrosine Natural Compound (not approved); | HDAC III (SIRT1) | Inhibits HDAC activity via the coordination of zinc ion in catalytic pocket of HDAC with sulfhydryl group activated by a reducing agent. | Ovarian Cancer cells, Colon Cancer cells and Cervical Cancer cells | Displays significant cytotoxic activity, inhibits cell proliferation and upregulates expression of tumor-suppressor gene gelsolin in a dose-dependent manner | (207, 208) | |
| Endometrial Cancer cells | Inhibits cell proliferation, significantly induces H3 and H4 acetylation, upregulates expression of cyclin-dependent kinase inhibitor, p21, and downregulates expression of pRb, cyclins, and CDKs, promoting cell cycle arrest. | (209) | |||||
| Breast Cancer cells | Inhibits proliferation induces cell cycle arrest at G2/M and reduces SIRT1 activity protein expression levels and reduces nuclear SIRT1 levels. Increases p53 acetylation (target of SIRT1) and increases DRAM expression | (208, 210) | |||||
| Fatty acid | Sodium Butyrate | Anti‐inflammatory | HDAC1 | Not described | Gastric Cancer cells | Increases DAPK expression in human gastric cancer cells and this expression prompted apoptosis by decreasing FAK levels. Suggesting that DAPK expression prompts apoptosis by reducing the FAK protein level. Induce demethylation of the SFRP gene promoter | (211) |
| Breast Cancer cells | Decreases cell proliferation induces cell cycle arrest at G1/G2 and decreases nuclear expression of DNA DSB repair proteins induced by etoposide (BRCA1 RAD51, ATM). Also increases H4 acetylation. | (212) | |||||
| Prostate Cancer cells | Inhibits HDAC1, 3 activity and induces H3, H4 acetylation, leading to hyperacetylation of H3 and H4 on the p21 promoter region, thus increasing p21 expression. Also induces cell cycle arrest, promoting apoptosis. | (213) | |||||
| Hydroxamic acid | Trichostatin A (TSA) | Antifungal | HDAC class I, II and SIRT6 | Hydroxamate. Pan-HDAC inhibitor, that obtain the binding energy associated with the strength of inhibition is derived from the bidentate chelation of hydroxamate | Breast Cancer cells | Decreases cell proliferation, inhibits HDAC activity, thus increases H4 hyperacetylation And increases ER acetylation and anti-tumor activity | (214) |
| Myeloid Leukemia (AML) cells | Inhibits HDAC activity leds to histone hyperacetylation. Increases H4 acetylation and reduces Myc expression ZNF278 (Myc’s coactivator), NM1, HOXB6 and MKRN3 | (215) | |||||
| Esophageal Squamous Carcinoma cells | Decreases cell proliferation, induces cell cycle arrest at G1, down-regulates cell growth by inhibiting the activation of the PI3K/Akt and ERK1/2 pathways, and increases H4 acetylation levels |
(216) | |||||
| Prostate Cancer cells | Increases apoptosis induces p21 expression and represses TMPRSS2-ERG expression AND affect acetylation status of p53 by inhibiting HDAC activity. Disrupts the epidermal growth factor receptor (EGFR),-STAT3 pathway, thus, inhibits proliferation in CRPC cells. Increases H4K16acetylation and promotes gene transcription, moreover decreases phospho-Akt pathway | (217, 218) | |||||
| Pancreatic Cancer cells | Restores cellular differentiation, reduces proliferation and restores p21 expression. Increases NDGR1 mRNA expression, also increases hypoxic responses | (219) | |||||
| Colon Cancer cells | Decreases cell growth and promotes apoptosis, down-regulates DNMT1 and HDAC1 expression, increasing p21, p27 and p57 expression | (220) | |||||
| Hepatocellular Carcinoma cells | Increases H3K9 and H3K27 acetylation and increases SERCA3 mRNA expression levels and promote ATP2A3 gene expression | (221) | |||||
| Vorinostat (SAHA) | Psoriasis disease treatmenr | HDAC class I, II and IV | inhibits HDAC activity by binding to the pocket of the catalytic site processes by removing acetyl groups from proteins | Advanced Prostate Cancer treatment | In a phase II trial, it was associated with significant toxicities limiting efficacy assessment in patients with disease progression on one prior chemotherapy | (222, 223) | |
| Follicular and Mantle Cell Lymphoma treatment | In a phase I trial in follicular and mantle cell lymphomaoral vorinostat was well tolerated up to 200mg bd for 14 consecutive days every 3 weeks in Japanese patients with NHL. Shown favorable results | (224) | |||||
| Panobinostat | HDACi multiple myeloma | HDAC | Pan-HDAC inhibitor, blocks the enzymatic activity of HDAC | Multiple myeloma treatment | It has improved progression-free survival when combined with bortezomib and dexamethasone in patients with relapsed multiple myeloma who previously received bortezomib and an immunomodulatory agent | (225) | |
| Depsipeptide | Burkholdacs A | Pathogen bacteria (not approved) | HDAC1, HDAC6 | Inhibits HDAC catalytic activity by reduction of the disulfide bond which generates a free thiol group that interacts with the catalytic site in HDACs. | Brain Cancer cells | In at least six cancer cell lines, it has shown superior HDACi activity over Ramidopsine (approved HDACi). Burkholdacs A presents more affinity for HDAC1 and was determined to be superior than B with respect to its HDAC1 inhibitory activity and isoform selectivity toward HDAC1 over HDAC6 and antiproliferative activity. | (226) |
| Colon Cancer cells | |||||||
| Lung Cancer cells | |||||||
| Ovary Cancer cells | |||||||
| Stomach Cancer cells | |||||||
| Prostate Cancer cells | |||||||
| Thailandepsin (Burkholdacs B) | Pathogen bacteria (not approved) | HDAC6 | Cervical Cancer cells | (226, 227) | |||
| Breast Cancer cells | (227) | ||||||
| Spiruchostatin A | Pathogen bacteria (not approved) | HDAC1 and HDAC6 | Structural similarity with HDAC inhibitor FK228 (Romidepsin) which interacts with the active-site zinc in its reduced form, preventing it from interacting with substrate. | Breast Cancer cells | Increases acetylation levels of specific lysine residues of histones H3 and H4. | (228, 229) | |
| Ovarian Cancer cells | |||||||
| Brain Cancer cells | |||||||
| Colon Cancer cells | |||||||
| Apicidin | Antiprotozoal for Malaria | HDAC3, HDAC4 and HDAC8 | Cyclic tripeptide that chelate the active site zinc ion through the terminal carbonyl, hydroxy and/or amino functional groups | Promyelocytic Leukemia treatment | Inhibits cell proliferation an cycle arrest, promoting cell death. Increases H4 acetylation and inhibits HDAC activity, thus increases p21 expression. | (230) | |
| Lung, Colon and Pancreatic Cancer cells | Induces DNA demethylation via HMT suppression, reduces HP1 and DNMT1 recruitment to genes’ promoter and induces p16, SALL3, and GATA4 expression. Also, decreases SUV39 and G9a expression in lung cancer cell lines. | (231) | |||||
| Cervical Cancer cells | Induces demethylation of CpG islands of the 1st exon of the PDH2 gene AND induces PHD2 and p21 gene expression and inhibits cell proliferation. | (232) | |||||
| Breast Cancer cells | Increases H3 and H4 acetylation and reduces ERalfa and ERb expression. Increases p21 and p27 expression and reduces cyclin D1 and cyclin E expression. Also reduces cell proliferation, thus promotig apoptosis. | (233) | |||||
| Endometrial Cancer cells | Increases H3 acetylation and reduces HDAC3, 4 expression, decreases cell proliferation and induces apoptosis. | (217) | |||||
| Ovarian Cancer cells | Decreases HDAC activity, reduces HDAC4 expression and blocks cell migration and invasion. Increases H3 and H4 acetylation and increases RECK expression through reducting the binding of HDAC4 to the Sp1 of its promoter, while reduces MMP-2 expression. | (234) | |||||
| Oral Squamous Cell Carcinoma cells | Inhibits cell growth, proliferation and reduces HDAC8 expression. Induces apoptosis and autophagy AND increases H4 acetylation. | (235) | |||||
| Platycodi | Nutraceutical (not approved) | HDAC | Not described | Lung Carcinoma cells | Inhibits HDAC enzymatic activity and induces the expression of p21. Stimulates cell death and inhibits cell proliferation. | (204) | |
Table 6
Current HAT HMT, HDM, and BET inhibitors repurposed with epigenetic applications in cancer therapy [*modified from Moreira-Silva et al., (9)].
ClassCompoundFirst indicationEpigenetic targetDrug-target interactionCancer model/New indicationKey features in mechanismReferences
| HAT, HMT, HDM, and BET inhibitors | |||||||
| HATi | Anacardic acid | Anti‐inflammatory; food-derived (not approved) | HAT/Ep300 and Tip60 | Not described | Cervical Tumor cells | Inhibits Tip60 HAT and ATM acetylation. Promotes resensitizing tumor cells to the cytotoxic effect of radiation. | (246) |
| Myeloid Leukemia cells | Inhibits p300 HAT activity. Also, inhibits NF-kB activation, inhibits IkBalfa activation, p65 acetylation and nuclear translocation. It potentiates apoptosis via TNF-induced caspase activation and suppresses the expression of genes involved in invasion and angiogenesis. | (247, 248) | |||||
| T‐Cell Lymphoma cells | |||||||
| Lung Cancer cells | |||||||
| Prostate Cancer cells | |||||||
| Garcinol | Antioxidant benzophenone (not approved); | HAT2B/Ep300 | Not described | Cervical Cancer cells | Inhibits p300 and KAT2B activity, HAT activity and induces apoptosis. | (249) | |
| Breast Cancer cells | Decreases H3K18 acetylation and increases DNA damage signaling markers. Inhibits HAT activity and induces cell proliferation arrest. | (250) | |||||
| Hepatocellular Carcinoma cells | Decreases HAT activity and inhibits STAT3 activation through acetylation. Decreases proliferation, tumor growth, survival and angiogenesis. | (251) | |||||
| Esophageal Carcinoma cells | Decreases p300/CBP levels, induces cell cycle arrest, thus induces apoptosis and inhibits migration and cell invasion and proliferation. Inhibits metastasis and inhibit HAT and its cofactors, decreasing TGF-beta pathway. | (252) | |||||
| Plumbagin | Nutraceutical quinone (not approved); | HAT3B/p300 | Inhibits p300 HAT activity (non-competitive), through a single hydroxyl group of plumbagin that makes a hydrogen bond with the lysine 1358 residue of the p300 HAT domain. |
Liver Carcinoma cells | Inhibits p300 HAT activity AND inhibits p300-mediated acetylation of p53 AND reduces H3 and H4 acetylation AND induces apoptosis AND modulates the enzymatic activity of p300. in vivo: reduces H3 acetylation. | (253) | |
| Lunasin | Natural Compound; food-derived peptide | HAT | Not described. Possibly a competitive inhibitor | Cancer preventive in mouse Fibroblasts | Suppresses foci formation in mice fibroblast cells induced by chemical carcinogens by the RGD motif and its chromatin-binding property, binding to deacetylated histones, and the reduction of histone acetylation. | (254) | |
| HMTi | Allantodapsone | Antibiotic (Dapsone-derivated) | H4R3me | Inhibitory activity toward PRMT1 | Hepatocellular Carcinoma cells | Inhibits cellular H4R3 methylation to the same level as AMI-1, while the H3K4 methylation level is barely impacted. | (255) |
| Ribavirin | RSV infections and Hepatitis C | EZH2 | Not described. Possibly a selective inhibitor of EZH2 | Solid Tumors (Atypical teratoid/rhabdoid tumor) | Inhibits cell growth, induces cell cycle arrest and apoptosis. Also inhibits eIF4E and EZH2 activity decreasing its expression levels. Impairs cell migration, invasion and adhesion. In osteosarcoma enhances chemosensitivity. | (230, 256) | |
| Breast, Brain, Cervical, Colon and Prostate Cancer cells | Decreases EZH2 expression, inhibits HMT activity and decreases H3K27me3. Induces variable growth inhibition and downregulation of EZH2, eIF4E and IMPDH1. | (257) | |||||
| Hydroxychloroquine (HCQ) | Antimalarial/Arthritis | PRC2 | Disruption of PRC2-EED complex by allosteric PRC2-EED binding inhibition within the H3K27me3-binding pocket, thus antagonizing the PRC2 catalytic activity | Multiple Myeloma Cells | Decreases H3K27me3 levels in MM cells 3 by disrupting the H3K27me3- EED interaction within the PRC2 complex. Suggesting that its anti-tumor activity might rely on the reactivation of genes abnormally silenced via H3K27 hypermethylation. | (258) | |
| HDMi | Clorgyline | MAO inhibitor | LSD1 | Not described | Bladder Cancer cells | Induces DNA demethylation, inhibits LSD1, decreasing H3K4me2 and H3K4me, establishes an active chromatin state. Inhibits cell growth induces the expression of previously silenced genes by enriching H3K4me2 and H3K4me1 histone marks. | (259) |
| Colon Cancer cells | |||||||
| Promyelocytic Leukemia Cells | |||||||
| Geranylgeranoic acid | Natural Compound (not approved) | LSD1 | Not described | Neuroblastoma cells | Inhibits LSD1 activity, induces NTRK2 gene expression and increases H3K4me2. Moreover decreases cell proliferation. | (260) | |
| Pargyline | MAO‐B inhibitor; antihypertensive | LSD1 | Not described | Prostate Cancer cells | Inhibits cell migration and invasion AND inhibit EMT AND induces E-cadherin expression AND inhibits N-cadherin and Vimentin expression AND delayed PCa transition to CRPC AND decreases PSA expression AND decreases H3K4 and H3K9 di-methylation. | (261) | |
| Tranylcypromine | Severe depression | LSD1 | Not described | Glioblastoma cells | Induces cell death AND inhibits LSD1 activity AND increases cell sensitivity to HDACi. | (262, 263) | |
| Polymyxin A/B | Antibiotic | LSD1 | Inhibits LSD1 by competition with its substrate at the enzyme’s cleft entry | Chemical inhibition of LSD1 assay | In vitro assays demonstrated that quinazoline core can represent a privileged scaffold for developing inhibitors that target epigenetic enzymes. | (264) | |
| BETi | Azelastine | Anti-histaminic | BET-BRD4 | Inhibits BRD4 through interactions with several key residues of the acetyl lysine binding pocket | Structural in silico assays by docking-based method | Docking-based database screening identified Azelastine drug as a promising novel template exhibiting binding affinity better than the control lead (+)-JQ1 for the human BRD4. Azelastine is having a low molecular weight, which gives a scope of further chemical modification to enrich its binding affinity for BRD4. |
(265) |
| Nitroxoline | Antibiotic | BET-BRD4 | Occupies the acetylated lysine binding pocket of the first bromodomain of BRD4 | MLL Leukemia cells | Prevents the binding of BRD4 to acetylated H4 | (266) | |
'암치료' 카테고리의 다른 글
| 승인된 약물의 용도 변경 (0) | 2021.08.22 |
|---|---|
| 종양학에서 약물 재배치 (0) | 2021.08.22 |
| 약물 용도 변경으로 암 치료 병목 극복 (0) | 2021.08.22 |
| 종양학의 용도 변경 약물: 후보 선택에서 임상 채택까지 (0) | 2021.08.22 |
| 암의 미토콘드리아 에너지 대사를 표적으로 하는 약물 용도 변경 (0) | 2021.08.22 |